Looking at your Data
2025-08-22
“The most certain sign of wisdom is cheerfulness.”
— Michel de Montaigne, Writer and philosopher
Plot Fonts and Theme
Code
library(systemfonts)
library(showtext)
## Clean the slate
systemfonts::clear_local_fonts()
systemfonts::clear_registry()
##
showtext_opts(dpi = 96) # set DPI for showtext
sysfonts::font_add(
family = "Alegreya",
regular = "../../../../../../fonts/Alegreya-Regular.ttf",
bold = "../../../../../../fonts/Alegreya-Bold.ttf",
italic = "../../../../../../fonts/Alegreya-Italic.ttf",
bolditalic = "../../../../../../fonts/Alegreya-BoldItalic.ttf"
)
sysfonts::font_add(
family = "Roboto Condensed",
regular = "../../../../../../fonts/RobotoCondensed-Regular.ttf",
bold = "../../../../../../fonts/RobotoCondensed-Bold.ttf",
italic = "../../../../../../fonts/RobotoCondensed-Italic.ttf",
bolditalic = "../../../../../../fonts/RobotoCondensed-BoldItalic.ttf"
)
showtext_auto(enable = TRUE) # enable showtext
##
theme_custom <- function() {
theme_bw(base_size = 10) +
#
# theme(panel.widths = unit(11, "cm"),
# panel.heights = unit(6.79, "cm")) + # Golden Ratio
theme(
plot.margin = margin_auto(t = 1, r = 2, b = 1, l = 1, unit = "cm"),
plot.background = element_rect(
fill = "bisque",
colour = "black",
linewidth = 1
)
) +
theme_sub_axis(
title = element_text(
family = "Roboto Condensed",
size = 10
),
text = element_text(
family = "Roboto Condensed",
size = 8
)
) +
theme_sub_legend(
text = element_text(
family = "Roboto Condensed",
size = 6
),
title = element_text(
family = "Alegreya",
size = 8
)
) +
theme_sub_plot(
title = element_text(
family = "Alegreya",
size = 14, face = "bold"
),
title.position = "plot",
subtitle = element_text(
family = "Alegreya",
size = 10
),
caption = element_text(
family = "Alegreya",
size = 6
),
caption.position = "plot"
)
}
## Use available fonts in ggplot text geoms too!
ggplot2::update_geom_defaults(geom = "text", new = list(
family = "Roboto Condensed",
face = "plain",
size = 3.5,
color = "#2b2b2b"
))
ggplot2::update_geom_defaults(geom = "label", new = list(
family = "Roboto Condensed",
face = "plain",
size = 3.5,
color = "#2b2b2b"
))
ggplot2::update_geom_defaults(geom = "marquee", new = list(
family = "Roboto Condensed",
face = "plain",
size = 3.5,
color = "#2b2b2b"
))
ggplot2::update_geom_defaults(geom = "text_repel", new = list(
family = "Roboto Condensed",
face = "plain",
size = 3.5,
color = "#2b2b2b"
))
ggplot2::update_geom_defaults(geom = "label_repel", new = list(
family = "Roboto Condensed",
face = "plain",
size = 3.5,
color = "#2b2b2b"
))
## Set the theme
ggplot2::theme_set(new = theme_custom())
## tinytable options
options("tinytable_tt_digits" = 2)
options("tinytable_format_num_fmt" = "significant_cell")
options(tinytable_html_mathjax = TRUE)
## Set defaults for flextable
flextable::set_flextable_defaults(font.family = "Roboto Condensed")
We spoke of Experiments and Data Gathering in the first module Nature of Data. This helped us to obtain data.
Our first task is to get acquainted with our data, to check the variables, the size of the dataset, how it is formatted, to eat it, as it were.
We need to inspect the data, to understand what it is telling us. The physical significance of each variable needs to sink in before we can do anything with it.
This is especially important in design, since we may be working in domains that are not within our own range of acqaintance or expertise. It is also an important step in the data analysis process.
What do we look for?
- How big is the dataset? How many rows and how many columns? Recall: Rows are observations, and columns are variables in the data
- What types of columns do we have? Quant? Qual? How many of each?
- What are the variable
names()? Are they adequate and memorable? - Is there missing data?
All this inspection will lead to:
- Data Cleaning, or Munging
- A clean dataset, whose variables we understand the meaning of, and which we will explore all the charts at our disposal.
Steps in Data Inspection and Cleaning
Inspection:
- Use
readr::read_csv()orreadr::read_delim()to read the data - Inspect Variables Names:
base::names()anddplyr::glimpse() - Discover Data Dimension/Size:
base::dim() - Structure of the data:
utils::str()[ Optional, but very useful. ] - Look for missing data:
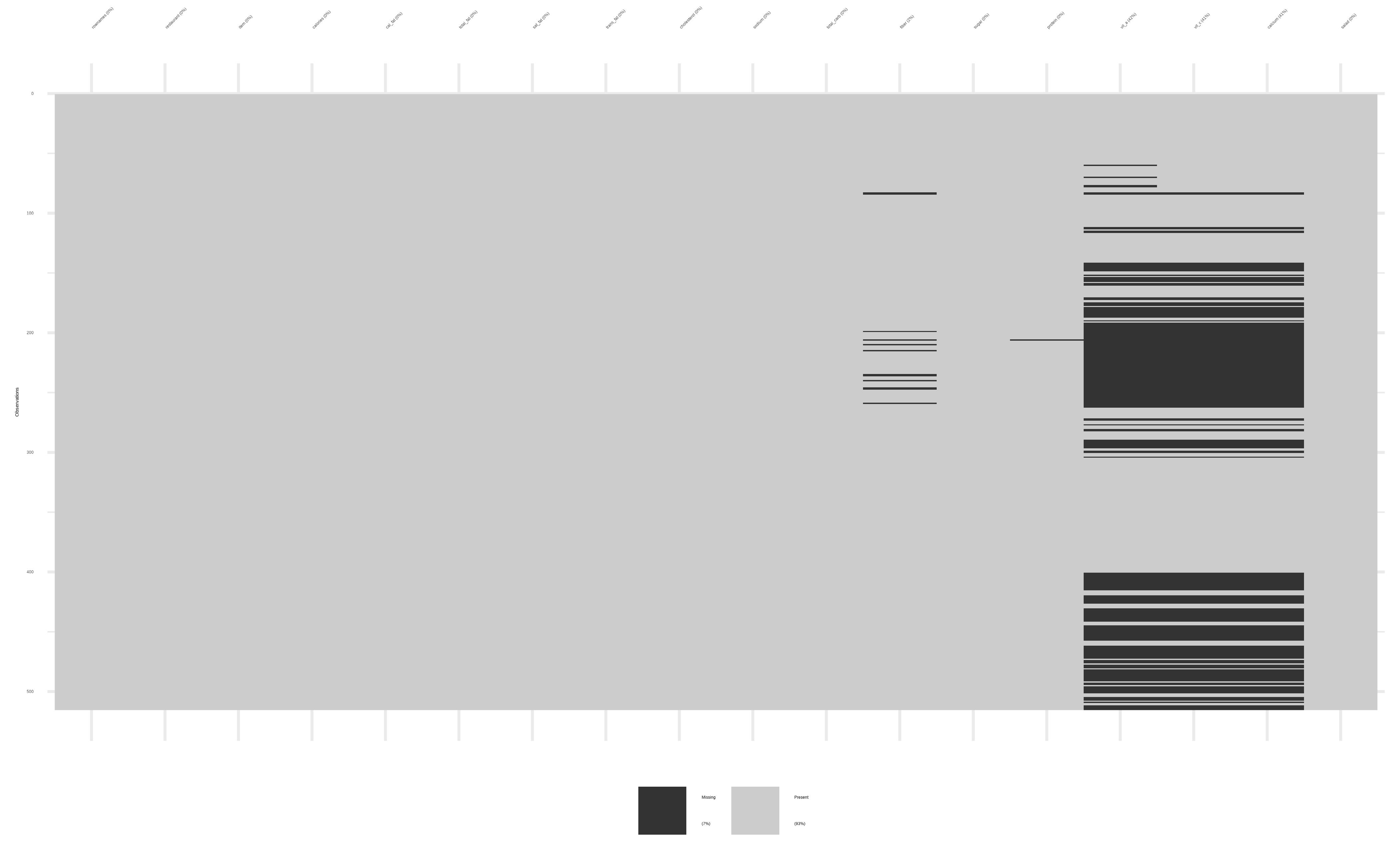
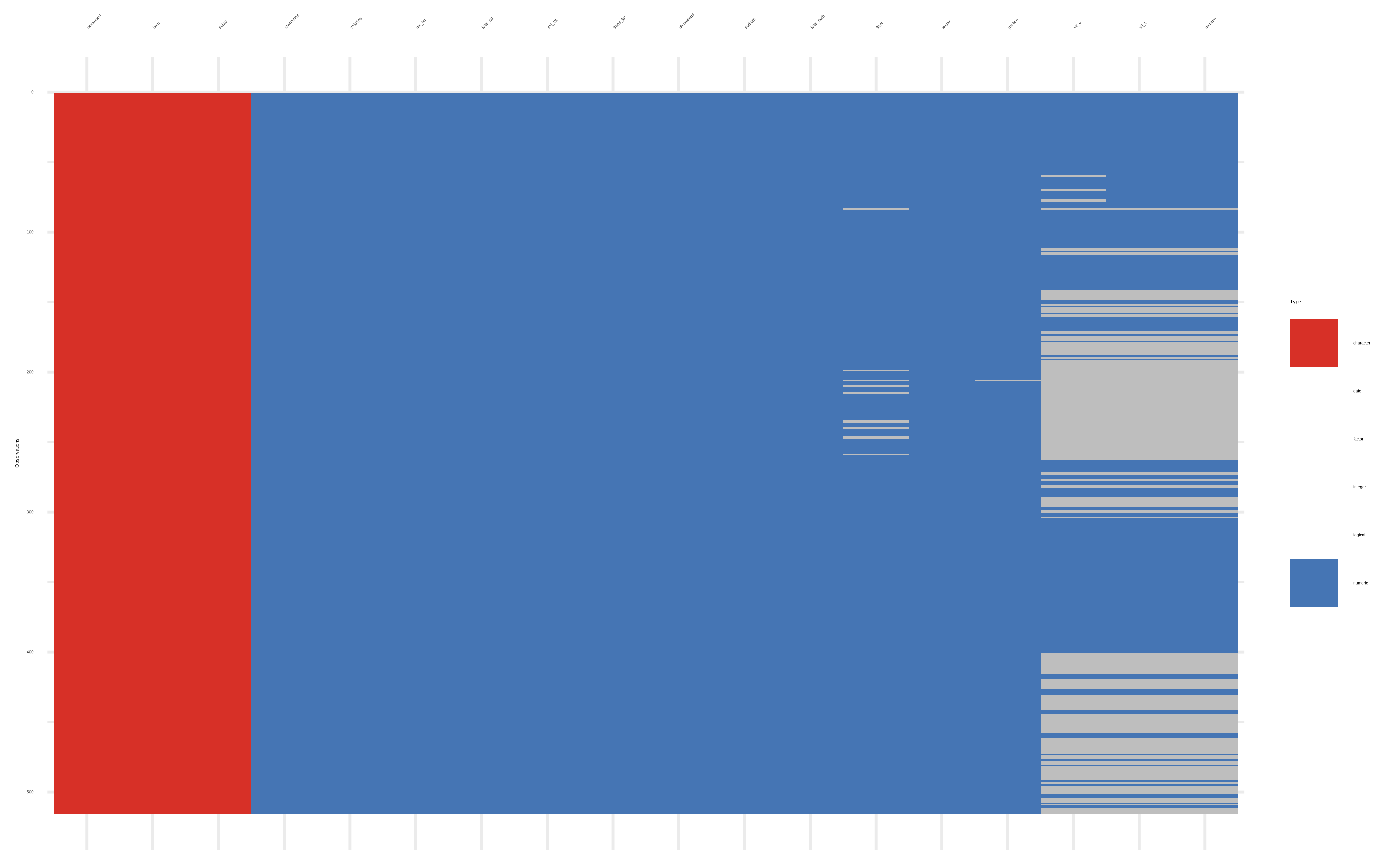
visdat::vis_dat()andvisdat::vis_miss()
And Munging:
- Clean the variable names:
janitor::clean_names() - Clean up missing data:
naniar::replace_with_na_all() - Make factors and rearrange factors to the left of our table using
dplyr::mutate(),base::factor()orforcats::as_factor(), followed bydplyr::relocate() - Make a cool table for our cleaned data with
tinytable::tt()(static) orDT::datatable()(interactive)
Since we are about to eat our data, we may begin with the dataset fastfood from the TidyTuesday Project for September 4, 2018.
Rows: 515 Columns: 18
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (3): restaurant, item, salad
dbl (15): rownames, calories, cal_fat, total_fat, sat_fat, trans_fat, choles...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.The output from readr::read_csv() tells us that the data frame contains 515 rows and 18 columns. At this point we can’t tell if there are missing values anywhere, or even if there are badly formatted data values anywhere.
Data Inspection
We already discover from the read_csv() output that the dataset has 515 rows and 18 columns. We can also use dim() to get this information:
This tells us that the dataset has 515 rows and 18 columns.
Again, read_csv() tells us that some columns are character, some are double and some are integer. We can use names() and dplyr::glimpse() to get more information about the variables in the dataset.
Rows: 515
Columns: 18
$ rownames <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,…
$ restaurant <chr> "Mcdonalds", "Mcdonalds", "Mcdonalds", "Mcdonalds", "Mcdon…
$ item <chr> "Artisan Grilled Chicken Sandwich", "Single Bacon Smokehou…
$ calories <dbl> 380, 840, 1130, 750, 920, 540, 300, 510, 430, 770, 380, 62…
$ cal_fat <dbl> 60, 410, 600, 280, 410, 250, 100, 210, 190, 400, 170, 300,…
$ total_fat <dbl> 7, 45, 67, 31, 45, 28, 12, 24, 21, 45, 18, 34, 20, 34, 8, …
$ sat_fat <dbl> 2.0, 17.0, 27.0, 10.0, 12.0, 10.0, 5.0, 4.0, 11.0, 21.0, 4…
$ trans_fat <dbl> 0.0, 1.5, 3.0, 0.5, 0.5, 1.0, 0.5, 0.0, 1.0, 2.5, 0.0, 1.5…
$ cholesterol <dbl> 95, 130, 220, 155, 120, 80, 40, 65, 85, 175, 40, 95, 125, …
$ sodium <dbl> 1110, 1580, 1920, 1940, 1980, 950, 680, 1040, 1040, 1290, …
$ total_carb <dbl> 44, 62, 63, 62, 81, 46, 33, 49, 35, 42, 38, 48, 48, 67, 31…
$ fiber <dbl> 3, 2, 3, 2, 4, 3, 2, 3, 2, 3, 2, 3, 3, 5, 2, 2, 3, 3, 5, 2…
$ sugar <dbl> 11, 18, 18, 18, 18, 9, 7, 6, 7, 10, 5, 11, 11, 11, 6, 3, 1…
$ protein <dbl> 37, 46, 70, 55, 46, 25, 15, 25, 25, 51, 15, 32, 42, 33, 13…
$ vit_a <dbl> 4, 6, 10, 6, 6, 10, 10, 0, 20, 20, 2, 10, 10, 10, 2, 4, 6,…
$ vit_c <dbl> 20, 20, 20, 25, 20, 2, 2, 4, 4, 6, 0, 10, 20, 15, 2, 6, 15…
$ calcium <dbl> 20, 20, 50, 20, 20, 15, 10, 2, 15, 20, 15, 35, 35, 35, 4, …
$ salad <chr> "Other", "Other", "Other", "Other", "Other", "Other", "Oth…By and large, the entries look good. There are no cases, immediately, of character data lurking in Quant variables and such like.
Data Munging
We need to deal with:
- Variable Naming
- Variable Type conversion
- Dealing with Missing Data
- Locating Variables for Attention!
As a part of the process, we should make sure that the variable names (not entries!!) are formatted in a “clean” way: there are a few options here, such as camelCase, snake_case, kebab-case, or dot.case. We will use the {janitor} package to do this, and also to make sure that the variable names are unique. AND, we will stick with snake_case for the rest of this course.
In this specific case, the variables names look evocative and meaningful enough, without being verbose; they seem just right. But as names in data become complex, with special characters ( %$#@!*_|? etc.), this becomes very useful.
We will also not touch the original data, but save the modified data in a new variable called fast_food_modified. This is a good practice, as it allows us to keep the original data intact, and also to compare the two if needed.
Name Cleaning
This cleaning up was not needed here, since the original names were already good. But it is a good practice to do this, as it will save you a lot of headaches later on.
Let us use the {visdat} package to visualize this:
Code
What to Do with Missing Data
It is clear that there are quite a few missing values in a few columns: vit_a, vit_c and calcium. Some missing values are also present in fiber. So what can one do?
A. Remove rows with missing values: We can use the tidyr::drop_na() command to check for empty locations in a any column, and drop rows containing NA values. Note that this will remove entire rows with missing values in any column, keeping only complete rows. This is a drastic step, and should be done with care.
B. Impute missing values: “Imputation” refers to a technique of inserting data values where they are lacking. This is for a more sophisticated data practitioner, and also requires domain expertise into the subject matter of the dataset itself. We can use the simputation package to impute missing values using various methods, such as trend detection for Quant variables, and using classification for Qual data. This is a more advanced topic, and we will not cover it here.
For our work here, to learn, we will use method A, and simply drop the cells containing NA, whenever we have to.
The {naniar} package
Missing or NA values can also, sadly, be encoded by humans using sets of characters or even default numbers! The {naniar} package has two built-in lists for common missing value codes:
naniar::common_na_numbersfor Quant variables, andnaniar::common_na_strings.
We can use these to replace these values with NA.
Replace Missing Values with NA
Code
Rows: 515
Columns: 18
$ rownames <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,…
$ restaurant <chr> "Mcdonalds", "Mcdonalds", "Mcdonalds", "Mcdonalds", "Mcdon…
$ item <chr> "Artisan Grilled Chicken Sandwich", "Single Bacon Smokehou…
$ calories <dbl> 380, 840, 1130, 750, 920, 540, 300, 510, 430, 770, 380, 62…
$ cal_fat <dbl> 60, 410, 600, 280, 410, 250, 100, 210, 190, 400, 170, 300,…
$ total_fat <dbl> 7, 45, 67, 31, 45, 28, 12, 24, 21, 45, 18, 34, 20, 34, 8, …
$ sat_fat <dbl> 2.0, 17.0, 27.0, 10.0, 12.0, 10.0, 5.0, 4.0, 11.0, 21.0, 4…
$ trans_fat <dbl> 0.0, 1.5, 3.0, 0.5, 0.5, 1.0, 0.5, 0.0, 1.0, 2.5, 0.0, 1.5…
$ cholesterol <dbl> 95, 130, 220, 155, 120, 80, 40, 65, 85, 175, 40, 95, 125, …
$ sodium <dbl> 1110, 1580, 1920, 1940, 1980, 950, 680, 1040, 1040, 1290, …
$ total_carb <dbl> 44, 62, 63, 62, 81, 46, 33, 49, 35, 42, 38, 48, 48, 67, 31…
$ fiber <dbl> 3, 2, 3, 2, 4, 3, 2, 3, 2, 3, 2, 3, 3, 5, 2, 2, 3, 3, 5, 2…
$ sugar <dbl> 11, 18, 18, 18, 18, 9, 7, 6, 7, 10, 5, 11, 11, 11, 6, 3, 1…
$ protein <dbl> 37, 46, 70, 55, 46, 25, 15, 25, 25, 51, 15, 32, 42, 33, 13…
$ vit_a <dbl> 4, 6, 10, 6, 6, 10, 10, 0, 20, 20, 2, 10, 10, 10, 2, 4, 6,…
$ vit_c <dbl> 20, 20, 20, 25, 20, 2, 2, 4, 4, 6, 0, 10, 20, 15, 2, 6, 15…
$ calcium <dbl> 20, 20, 50, 20, 20, 15, 10, 2, 15, 20, 15, 35, 35, 35, 4, …
$ salad <chr> "Other", "Other", "Other", "Other", "Other", "Other", "Oth…Note that with large datasets, this replacement of strings and numbers with naniar::replace_with_na_all() takes a lot of time to execute.
We see that there are certain variables that must be converted to factors for analytics purposes, since they are unmistakably Qualitative in nature. Let us do that now, for use later:
Code
fast_food_modified <- fast_food_modified %>%
mutate(
restaurant = as.factor(restaurant),
salad = as.factor(salad),
item = as.factor(item)
) %>%
rename("dish" = item) %>% # rename item to dish
# arrange the Qual variables first, Quant next
dplyr::relocate(where(is.factor), .after = rownames)
glimpse(fast_food_modified)Rows: 515
Columns: 18
$ rownames <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,…
$ restaurant <fct> Mcdonalds, Mcdonalds, Mcdonalds, Mcdonalds, Mcdonalds, Mcd…
$ dish <fct> "Artisan Grilled Chicken Sandwich", "Single Bacon Smokehou…
$ salad <fct> Other, Other, Other, Other, Other, Other, Other, Other, Ot…
$ calories <dbl> 380, 840, 1130, 750, 920, 540, 300, 510, 430, 770, 380, 62…
$ cal_fat <dbl> 60, 410, 600, 280, 410, 250, 100, 210, 190, 400, 170, 300,…
$ total_fat <dbl> 7, 45, 67, 31, 45, 28, 12, 24, 21, 45, 18, 34, 20, 34, 8, …
$ sat_fat <dbl> 2.0, 17.0, 27.0, 10.0, 12.0, 10.0, 5.0, 4.0, 11.0, 21.0, 4…
$ trans_fat <dbl> 0.0, 1.5, 3.0, 0.5, 0.5, 1.0, 0.5, 0.0, 1.0, 2.5, 0.0, 1.5…
$ cholesterol <dbl> 95, 130, 220, 155, 120, 80, 40, 65, 85, 175, 40, 95, 125, …
$ sodium <dbl> 1110, 1580, 1920, 1940, 1980, 950, 680, 1040, 1040, 1290, …
$ total_carb <dbl> 44, 62, 63, 62, 81, 46, 33, 49, 35, 42, 38, 48, 48, 67, 31…
$ fiber <dbl> 3, 2, 3, 2, 4, 3, 2, 3, 2, 3, 2, 3, 3, 5, 2, 2, 3, 3, 5, 2…
$ sugar <dbl> 11, 18, 18, 18, 18, 9, 7, 6, 7, 10, 5, 11, 11, 11, 6, 3, 1…
$ protein <dbl> 37, 46, 70, 55, 46, 25, 15, 25, 25, 51, 15, 32, 42, 33, 13…
$ vit_a <dbl> 4, 6, 10, 6, 6, 10, 10, 0, 20, 20, 2, 10, 10, 10, 2, 4, 6,…
$ vit_c <dbl> 20, 20, 20, 25, 20, 2, 2, 4, 4, 6, 0, 10, 20, 15, 2, 6, 15…
$ calcium <dbl> 20, 20, 50, 20, 20, 15, 10, 2, 15, 20, 15, 35, 35, 35, 4, …Data Dictionary
Using all the above methods, we can now create a data dictionary for the fastfood_modified dataset. This is a good practice, as it helps us to understand the data better, and also to communicate with others about the data.
Quantitiative Data
calories(int): Calories in the dishcalories_from_fat(int): Calories from fattotal_fat_g(dbl): Total fat in gramssaturated_fat_g(dbl): Saturated fat in gramstrans_fat(dbl): Trans fat in gramscholesterol_mg(int): Cholesterol in milligramssodium_mg(int): Sodium in milligramscarbohydrates_g(dbl): Carbohydrates in gramsfiber_g(dbl): Fiber in gramssugars_g(dbl): Sugars in gramsprotein_g(dbl): Protein in gramsvit_a(int): Vitamin A in % Daily Valuevit_c(int): Vitamin C in % Daily Valuecalcium(int): Calcium in % Daily Valueiron(int): Iron in % Daily Value
Qualitative Data
restaurant(fct): Name of the restaurantitem(fct): Name of the dishsalad(fct): Is it a salad? (Yes/No)rownames(int): Row ID
It is usually a good idea to make crisp business-like tables to show your data. There are many methods to do this.
Static Table Reporting
For Static Tables ( to be published in reports, papers, etc.), one of the simplest and effective ones is to use the tt set of commands from {tinytable}. (The kable set of commands from the {knitr} and {kableExtra} packages also are a good choice):
Code
fast_food_modified %>%
head(10) %>%
tinytable::tt(caption = "Fast Food Dataset (Clean)") %>%
tinytable::theme_html(class = "table table-hover table-striped table-condensed") %>%
style_tt(fontsize = 0.8) %>%
stats::setNames(c("Row ID", "Restaurant", "Dish", "Calories", "Calories from fat", "Total Fat (g)", "Saturated Fat (g)", "Trans Fat (g)", "Cholesterol (mg)", "Sodium (mg)", "Carbohydrates (g)", "Fiber (g)", "Sugars (g)", "Protein (g)", "Vitamin A (% DV)", "Vitamin C (% DV)", "Calcium (% DV)", "Iron (% DV)"))| Row ID | Restaurant | Dish | Calories | Calories from fat | Total Fat (g) | Saturated Fat (g) | Trans Fat (g) | Cholesterol (mg) | Sodium (mg) | Carbohydrates (g) | Fiber (g) | Sugars (g) | Protein (g) | Vitamin A (% DV) | Vitamin C (% DV) | Calcium (% DV) | Iron (% DV) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mcdonalds | Artisan Grilled Chicken Sandwich | Other | 380 | 60 | 7 | 2 | 0 | 95 | 1110 | 44 | 3 | 11 | 37 | 4 | 20 | 20 |
| 2 | Mcdonalds | Single Bacon Smokehouse Burger | Other | 840 | 410 | 45 | 17 | 1.5 | 130 | 1580 | 62 | 2 | 18 | 46 | 6 | 20 | 20 |
| 3 | Mcdonalds | Double Bacon Smokehouse Burger | Other | 1130 | 600 | 67 | 27 | 3 | 220 | 1920 | 63 | 3 | 18 | 70 | 10 | 20 | 50 |
| 4 | Mcdonalds | Grilled Bacon Smokehouse Chicken Sandwich | Other | 750 | 280 | 31 | 10 | 0.5 | 155 | 1940 | 62 | 2 | 18 | 55 | 6 | 25 | 20 |
| 5 | Mcdonalds | Crispy Bacon Smokehouse Chicken Sandwich | Other | 920 | 410 | 45 | 12 | 0.5 | 120 | 1980 | 81 | 4 | 18 | 46 | 6 | 20 | 20 |
| 6 | Mcdonalds | Big Mac | Other | 540 | 250 | 28 | 10 | 1 | 80 | 950 | 46 | 3 | 9 | 25 | 10 | 2 | 15 |
| 7 | Mcdonalds | Cheeseburger | Other | 300 | 100 | 12 | 5 | 0.5 | 40 | 680 | 33 | 2 | 7 | 15 | 10 | 2 | 10 |
| 8 | Mcdonalds | Classic Chicken Sandwich | Other | 510 | 210 | 24 | 4 | 0 | 65 | 1040 | 49 | 3 | 6 | 25 | 0 | 4 | 2 |
| 9 | Mcdonalds | Double Cheeseburger | Other | 430 | 190 | 21 | 11 | 1 | 85 | 1040 | 35 | 2 | 7 | 25 | 20 | 4 | 15 |
| 10 | Mcdonalds | Double Quarter Pounder® with Cheese | Other | 770 | 400 | 45 | 21 | 2.5 | 175 | 1290 | 42 | 3 | 10 | 51 | 20 | 6 | 20 |
Interactive Table Reporting
Dynamic Tables can be easily made using the {DT} package, which allows for sorting, searching, and pagination. This is useful for exploring the data interactively. Here is an example:
Code
fast_food_modified %>%
DT::datatable(
style = "default",
caption = htmltools::tags$caption(
style = "caption-side: top; text-align: left; color: black; font-size: 100%;", "Fast Food Dataset (Clean)"
),
options = list(pageLength = 10, autoWidth = TRUE)
) %>%
DT::formatStyle(
columns = names(fast_food_modified),
fontFamily = "Roboto Condensed",
fontSize = "12px"
)
- See if you can do this for this messy dataset which you can download by clicking on the button below the table:
| species | island | bill_len | bill_dep | flipper_len | body_mass | sex | year |
|---|---|---|---|---|---|---|---|
| Adelie | 39.1 | 18.7 | 181 | 3750 | male | 2007 | |
| Adelie | Torgersen | 39.5 | 17.4 | 186 | 3800 | female | 2007 |
| Adelie | Torgersen | 40.3 | 18.0 | 195 | 3250 | female | 2007 |
| Adelie | Torgersen | NA | NA | NA | NA | NA | 2007 |
| Adelie | Torgersen | 999.0 | NA | 193 | 3450 | female | 2007 |
| Adelie | Torgersen | 39.3 | 20.6 | 190 | 3650 | male | 2007 |
Save it inside your data folder, and call it penguins_messy.csv. Then read the data in your Quarto document using readr::read_csv("data/penguins-messy.csv) and proceed.
- Install the package
{tastyR}. It contains two datasets,allrecipesandcuisines. Do a similar inspection and if needed, cleaning/munging of these datasets.
- Data Inspection is an essential step in getting to know your data.
- The structure and format of your data variables, what they mean, and what they might be telling you, is crucial to Exploring, Analysing, and Modelling with the data.
- Data Cleaning is an essential step in the data analysis process.
- These steps get much of the headache out of the way, and allow you to focus on the real work of Data Exploration, Data Analysis, and Modelling.
- And Data Presentation!!
- The first step in data analysis is to get to know your data.
- Use
readr::read_csv()to read the data. - Use
names(),glimpse(),dim(), andstr()to get to know the variables in your data. - Use
visdat::vis_miss()andvis_dat()to visualize missing data. - Use
naniar::replace_with_na_all()to replace missing values withNA. If it runs too slowly, then fall back totidyr::drop_na(). Strange looking strings, whichnaniarreplaces with ease, may have to be separately searched for and replaced, using a combination ofdplyr::mutate()andstr_detect(). - Use
janitor::clean_names()to clean the variable names. - Use
tinytable::tt()orDT::datatable()to create tables for your data.
Make these part of your Workflow.
- Nicholas Tierney. (2024-03-05). Getting Started with naniar. https://cran.r-project.org/web/packages/naniar/vignettes/getting-started-w-naniar.html
- Vincent Arel-Bundock. tinytable. https://vincentarelbundock.github.io/tinytable/.
- Vincent Arel-Bundock. RDatasets. https://vincentarelbundock.github.io/Rdatasets/.