Correlation and Regression Explorations
1 Packages
Plot Fonts and Theme
Show the Code
library(systemfonts)
library(showtext)
library(tidyverse)
## Clean the slate
systemfonts::clear_local_fonts()
systemfonts::clear_registry()
##
showtext_opts(dpi = 96) # set DPI for showtext
sysfonts::font_add(
family = "Alegreya",
regular = "../../../../../../../fonts/Alegreya-Regular.ttf",
bold = "../../../../../../../fonts/Alegreya-Bold.ttf",
italic = "../../../../../../../fonts/Alegreya-Italic.ttf",
bolditalic = "../../../../../../../fonts/Alegreya-BoldItalic.ttf"
)
sysfonts::font_add(
family = "Roboto Condensed",
regular = "../../../../../../../fonts/RobotoCondensed-Regular.ttf",
bold = "../../../../../../../fonts/RobotoCondensed-Bold.ttf",
italic = "../../../../../../../fonts/RobotoCondensed-Italic.ttf",
bolditalic = "../../../../../../../fonts/RobotoCondensed-BoldItalic.ttf"
)
showtext_auto(enable = TRUE) # enable showtext
##
theme_custom <- function() {
font <- "Alegreya" # assign font family up front
"%+replace%" <- ggplot2::"%+replace%" # nolint
ggplot2::theme_classic(base_size = 14, base_family = font) %+replace% # replace elements we want to change
theme(
text = element_text(family = font), # set base font family
# text elements
plot.title = element_text( # title
family = font, # set font family
size = 24, # set font size
face = "bold", # bold typeface
hjust = 0, # left align
margin = margin(t = 5, r = 0, b = 5, l = 0)
), # margin
plot.title.position = "plot",
plot.subtitle = element_text( # subtitle
family = font, # font family
size = 14, # font size
hjust = 0, # left align
margin = margin(t = 5, r = 0, b = 10, l = 0)
), # margin
plot.caption = element_text( # caption
family = font, # font family
size = 9, # font size
hjust = 1
), # right align
plot.caption.position = "plot", # right align
axis.title = element_text( # axis titles
family = "Roboto Condensed", # font family
size = 12
), # font size
axis.text = element_text( # axis text
family = "Roboto Condensed", # font family
size = 9
), # font size
axis.text.x = element_text( # margin for axis text
margin = margin(5, b = 10)
)
# since the legend often requires manual tweaking
# based on plot content, don't define it here
)
}
## Use available fonts in ggplot text geoms too!
ggplot2::update_geom_defaults(geom = "text", new = list(
family = "Roboto Condensed",
face = "plain",
size = 3.5,
color = "#2b2b2b"
))
## Set the theme
ggplot2::theme_set(new = theme_custom())2 Intro
I will work through and “unify” at least two things:
- Hadley Wickham’s chapter on modelling and his analysis of the linear model for the diamonds dataset
- The diagnostic aspects of Linear Regression as detailed in Crawley’s book
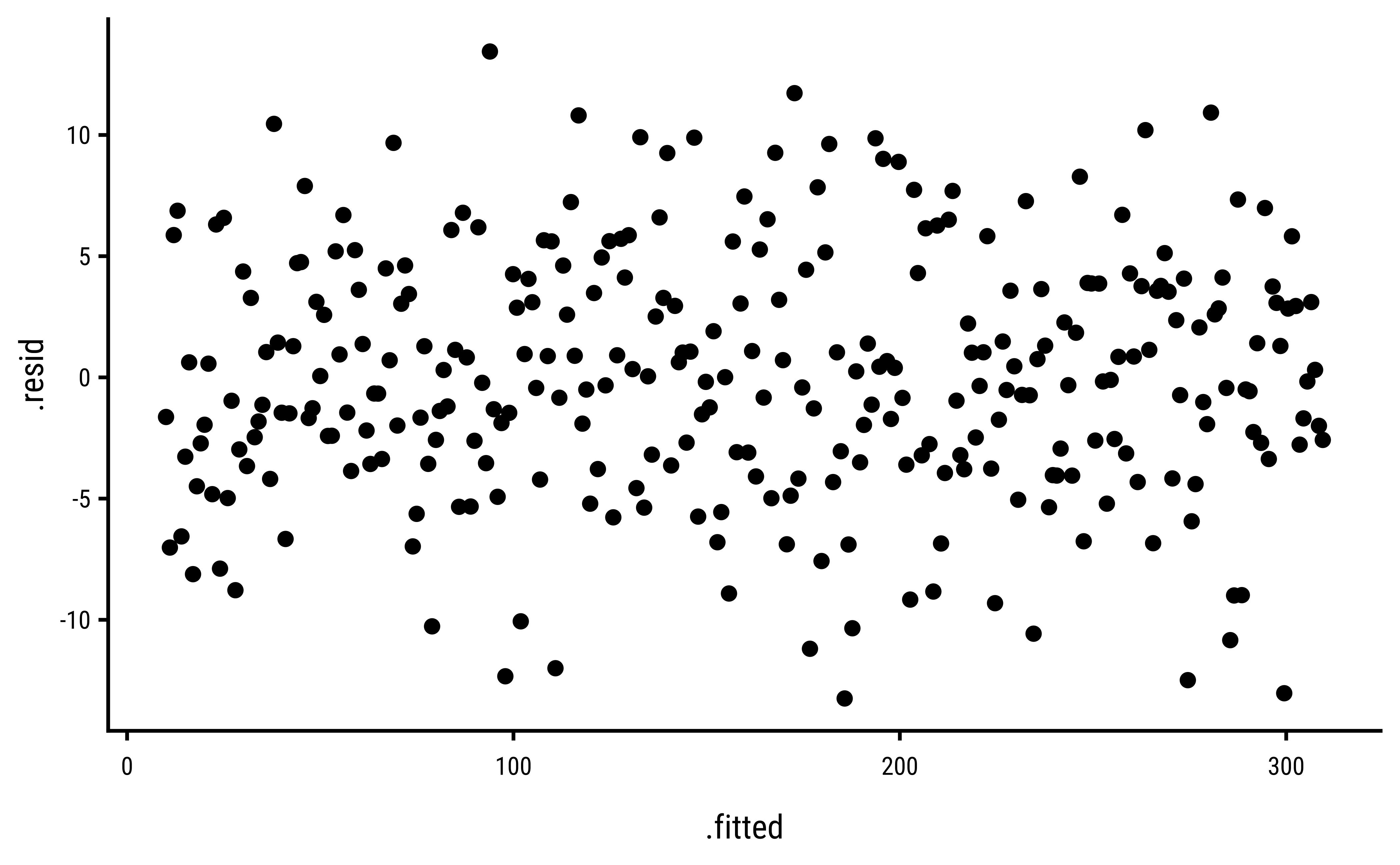
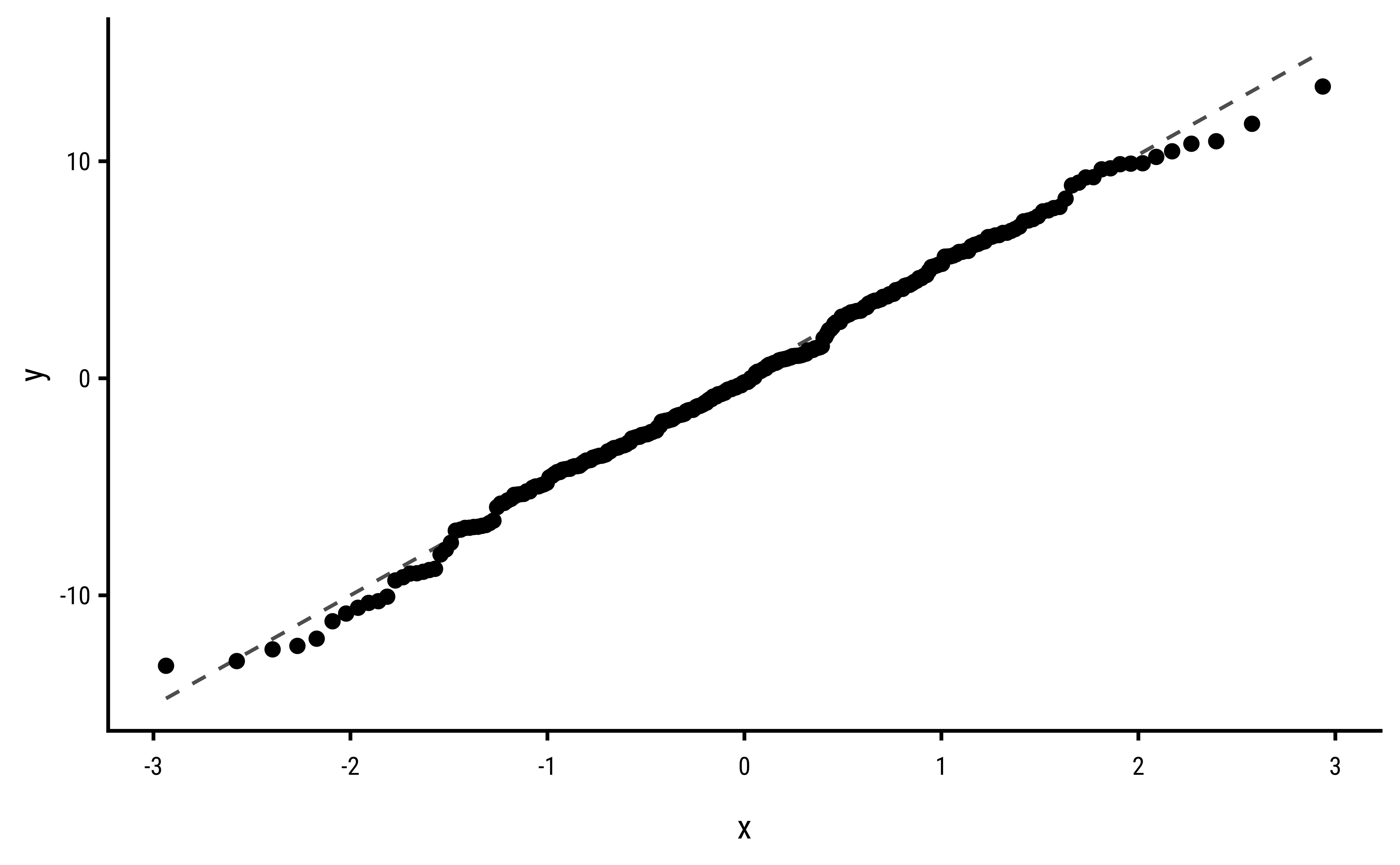
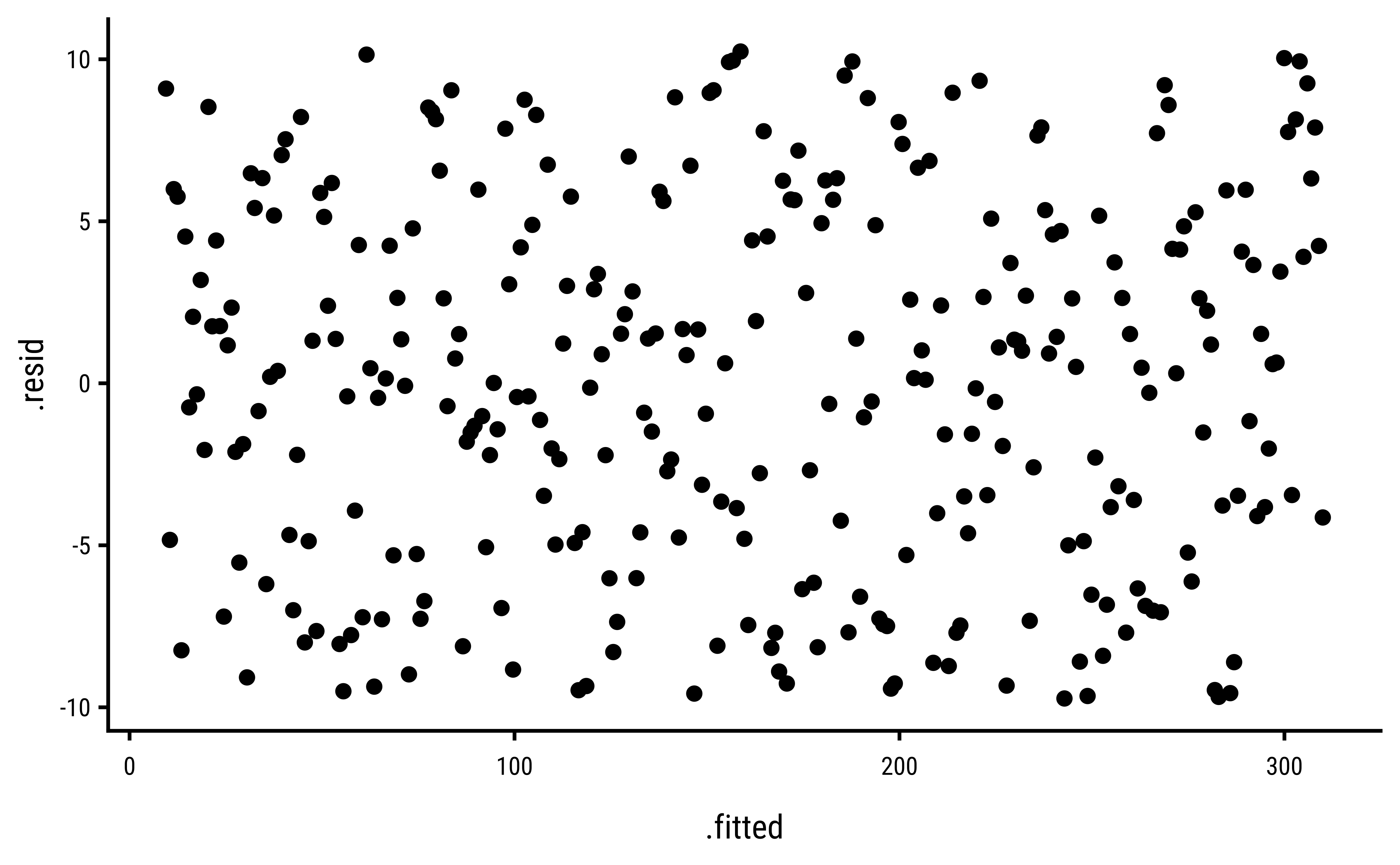
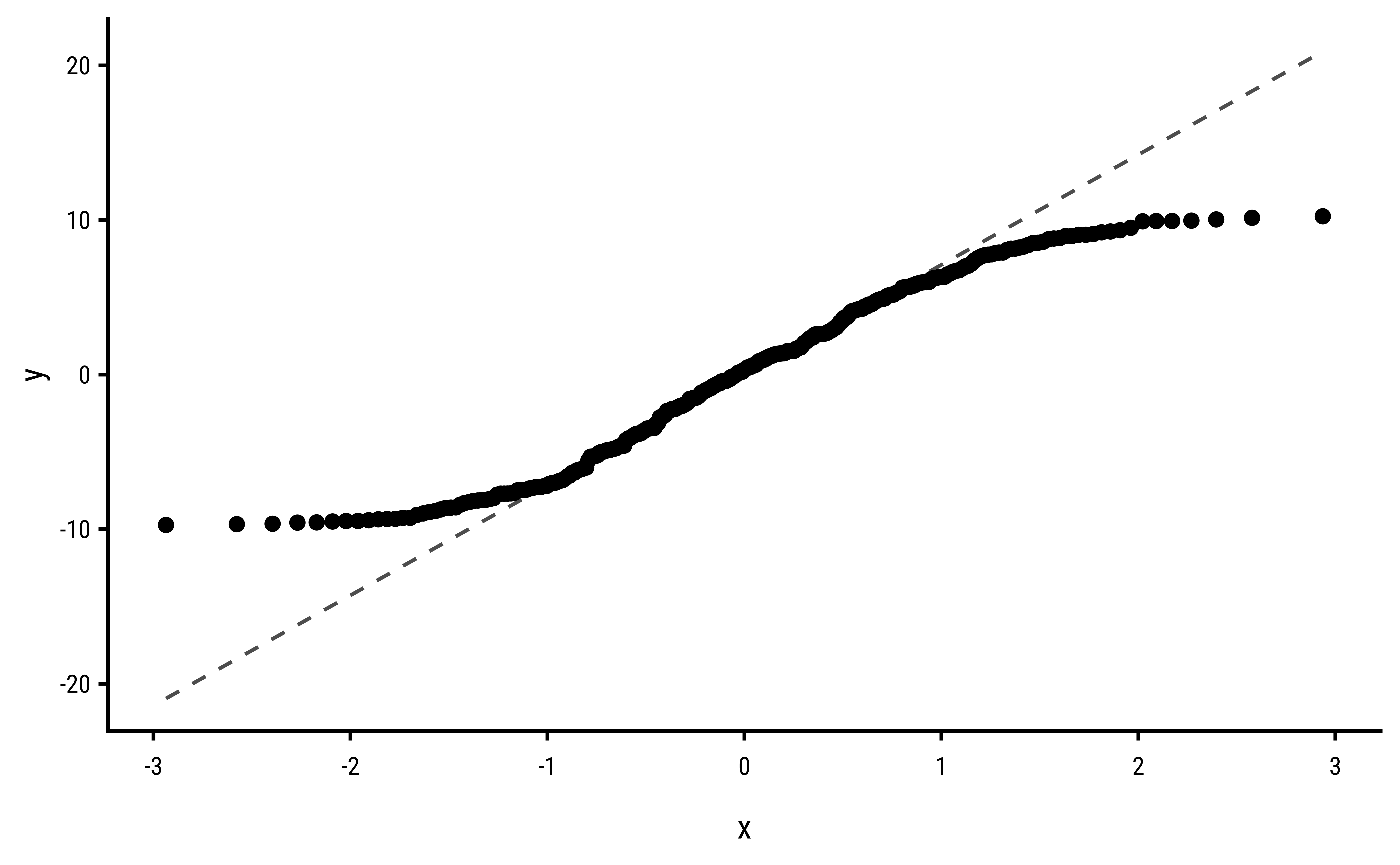
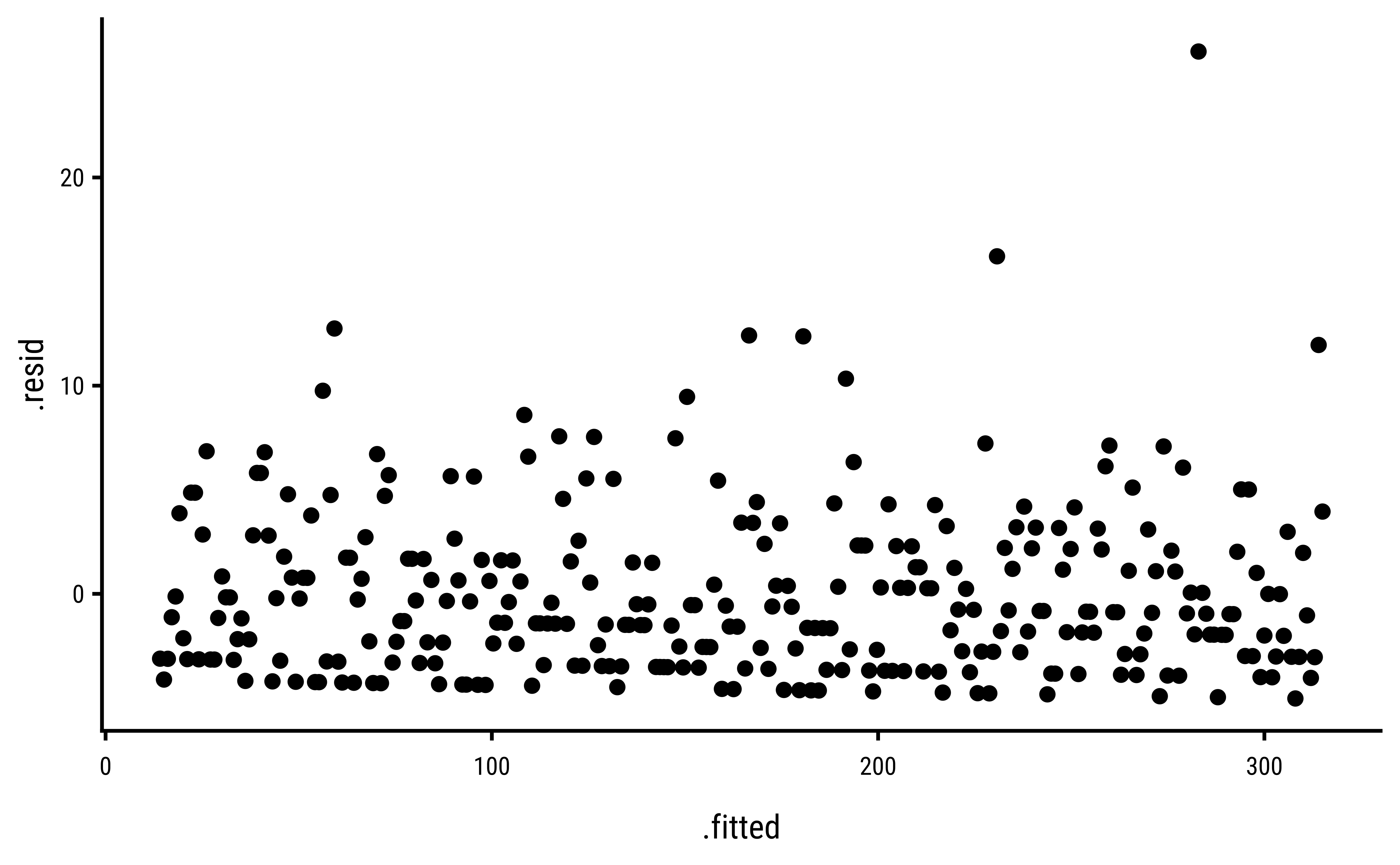
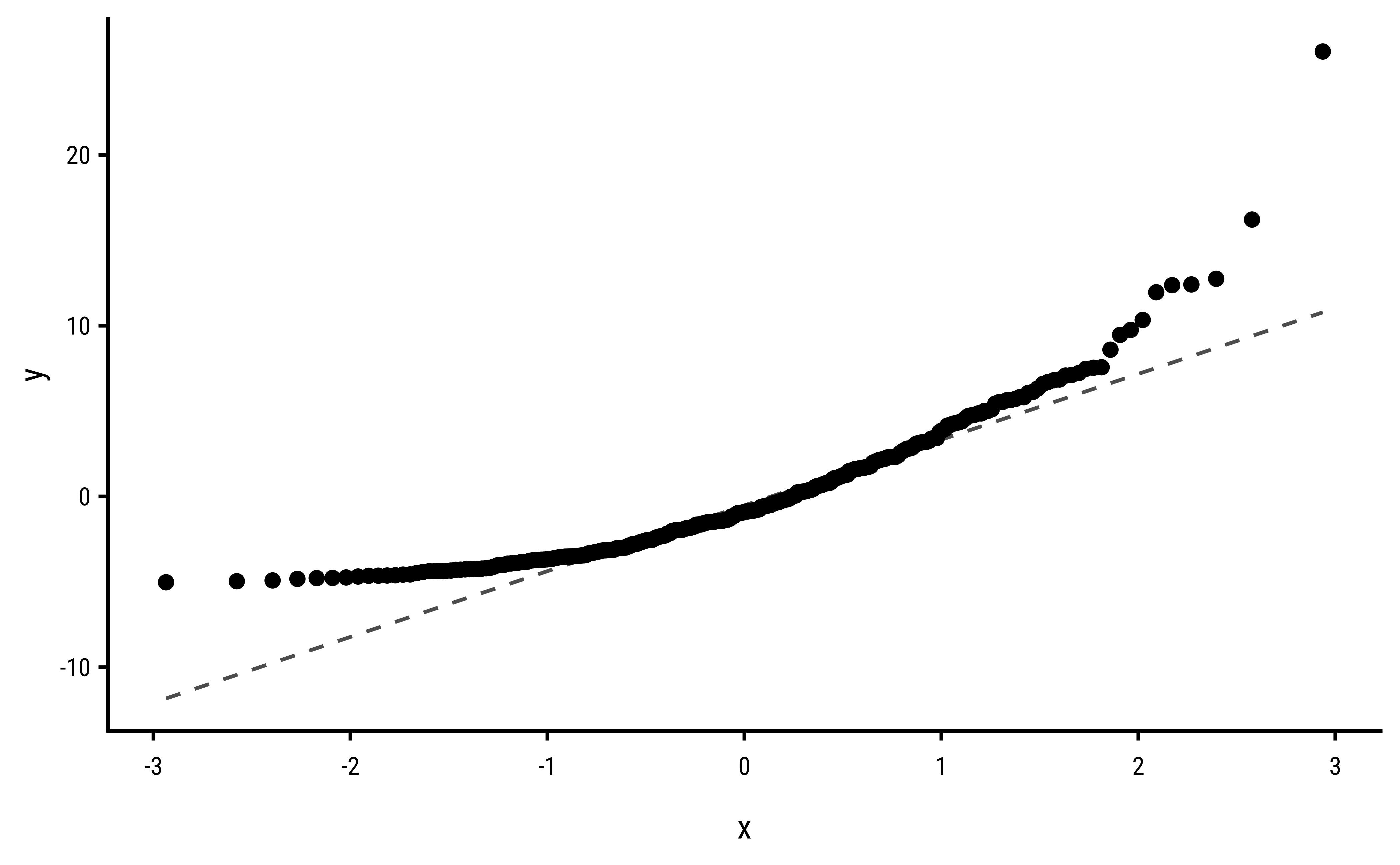
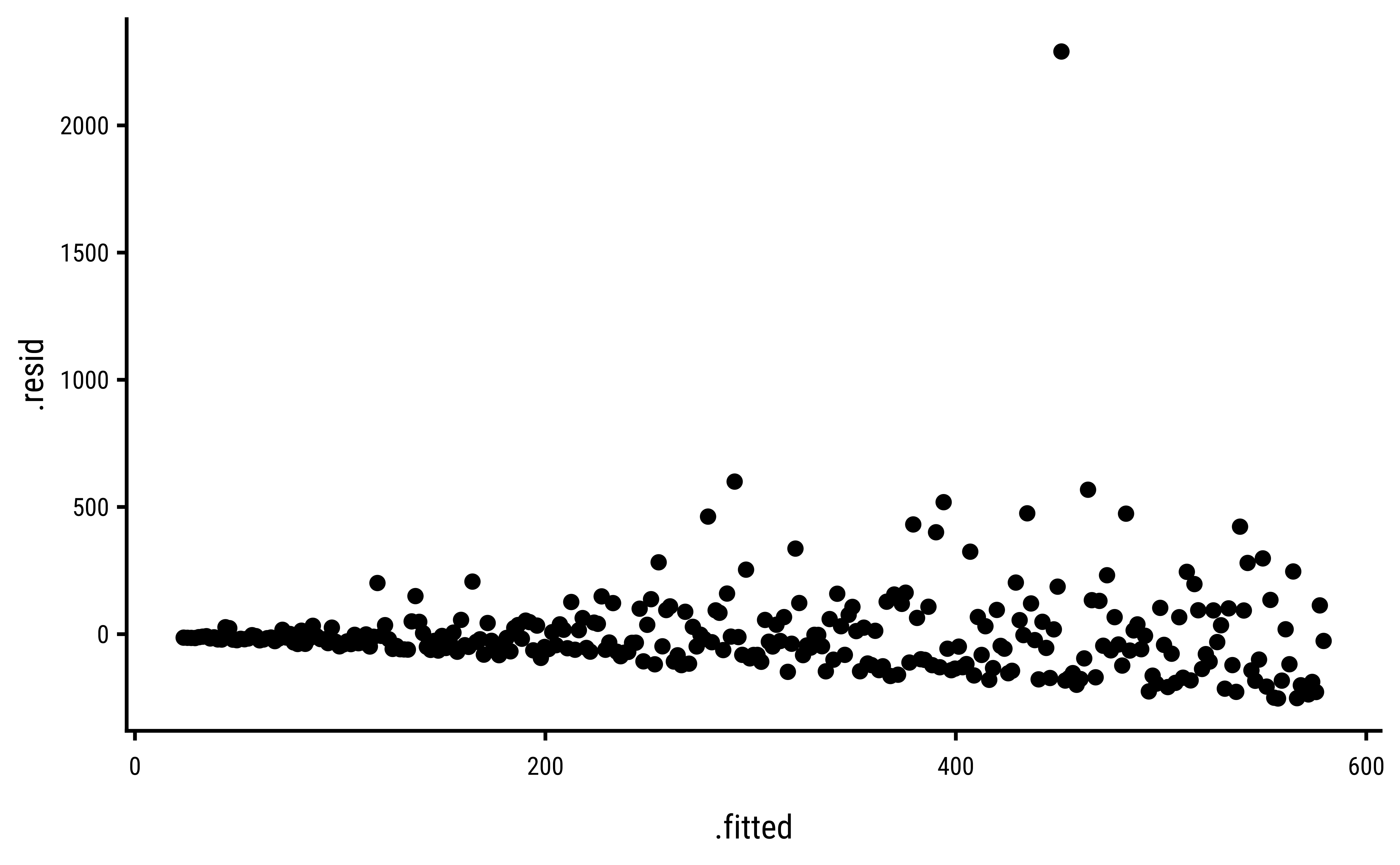
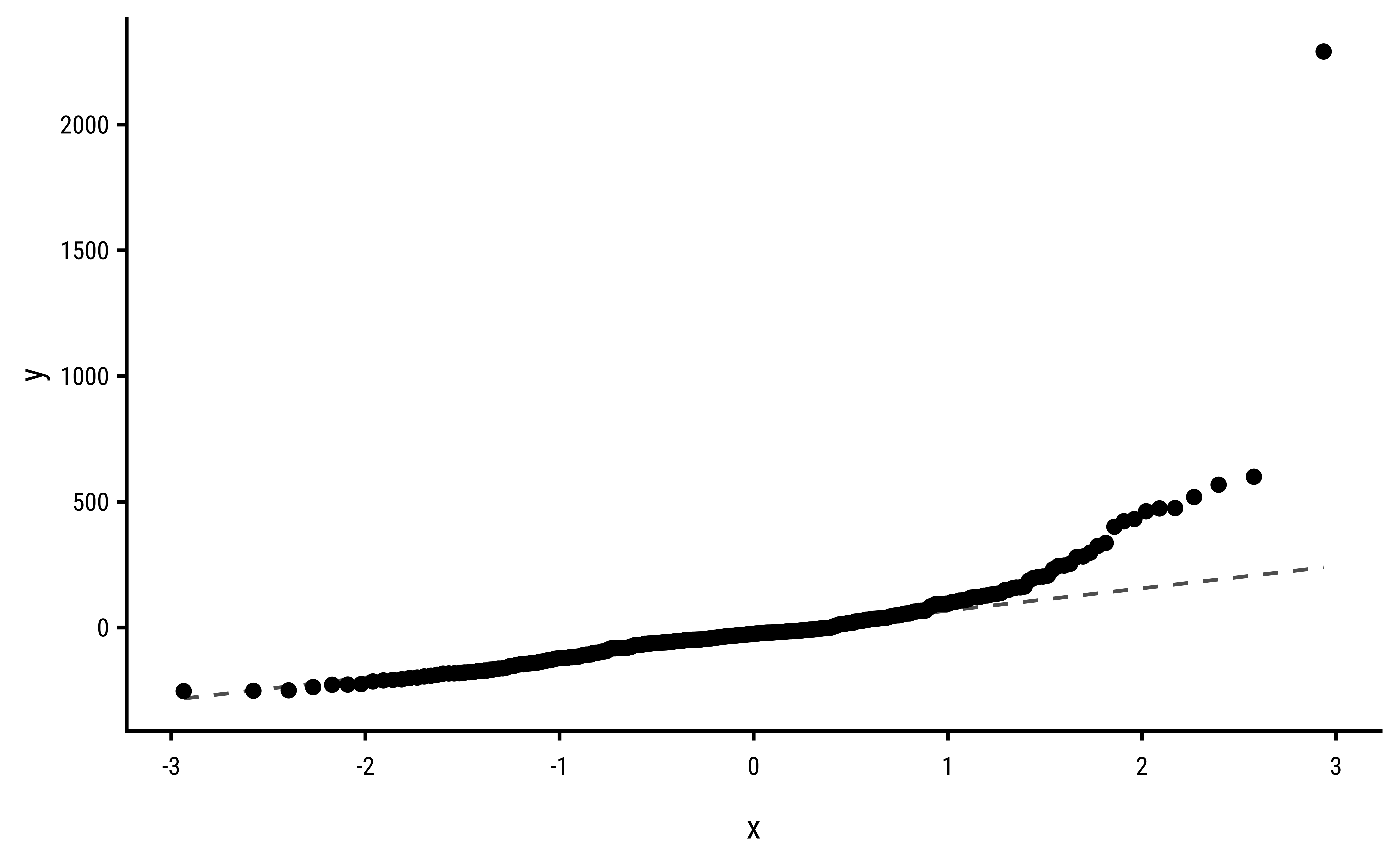
3 Explorations into Diagnostic Plots
Let us create dependent y* variables with different sorts of errors: